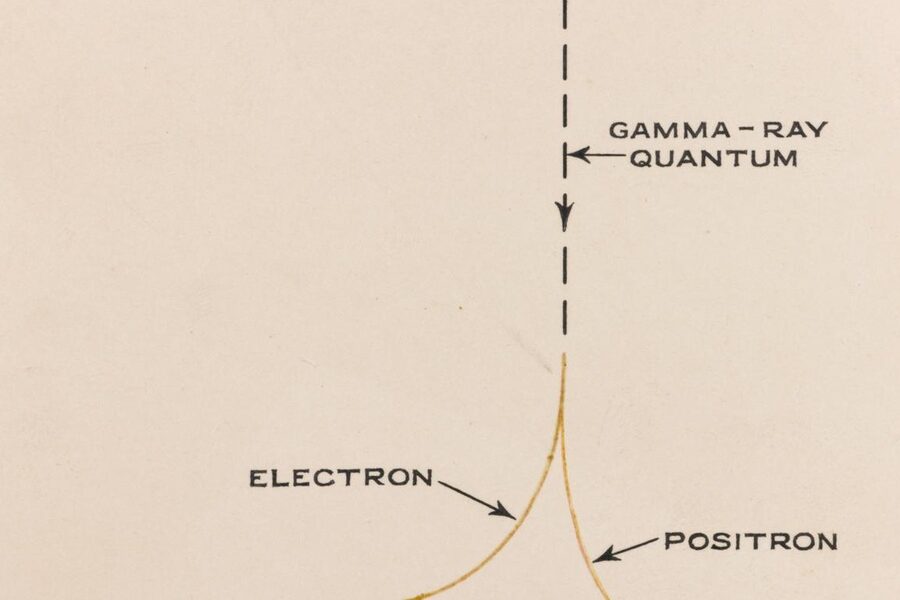
From living cells to engineered materials, the ways atoms stick together determine everything from protein folding to polymer strength. A quick grasp of bond types makes it easier to predict reactivity, stability, and function across chemistry and biology.
There are 22 Chemical Bonds, ranging from Amide (peptide) bond to π–π stacking. For each entry you’ll find below the columns Classification,Typical energy (kJ/mol),Common example so you can compare how bonds are grouped, their typical strengths, and representative cases at a glance — you’ll find the full list and details below.
How should I use the “Typical energy (kJ/mol)” values when comparing bonds?
Typical energy values are useful for relative comparisons: higher numbers usually mean stronger, harder-to-break interactions, but real strengths depend on context (solvent, geometry, neighboring groups). Treat the values as guidance for stability and reactivity trends rather than exact numbers for every situation.
Which bond types here matter most in biological systems?
In biology, covalent bonds (like the Amide/peptide bond) set primary structure, while hydrogen bonds, ionic interactions, disulfide bridges and π–π stacking shape folding and recognition; the list below highlights those and also weaker interactions that influence assembly and dynamics.
Chemical Bonds
| Name | Classification | Typical energy (kJ/mol) | Common example |
|---|---|---|---|
| Ionic bond | Primary ionic | 400-4,000 | Sodium chloride (NaCl) crystal |
| Metallic bond | Metallic | 100-800 | Copper metal (Cu) |
| Nonpolar covalent | Covalent nonpolar | 150-500 | H2, Cl2, C-H bonds |
| Polar covalent | Covalent polar | 200-600 | Hydrogen chloride (HCl) |
| Single covalent | Covalent single (sigma) | 150-400 | C–C single bond in ethane |
| Double covalent | Covalent double (sigma+pi) | 400-700 | C=C in ethene |
| Triple covalent | Covalent triple (sigma+2pi) | 600-900 | C≡C in acetylene |
| Coordinate (dative) bond | Covalent coordinate | 150-500 | NH3→BF3 adduct |
| Sigma bond (σ) | Covalent sigma | 200-700 | C–H in methane |
| Pi bond (π) | Covalent pi | 60-300 | C=C pi component in ethene |
| Aromatic bond (delocalized) | Covalent delocalized | 400-550 | Benzene ring (C6H6) |
| Disulfide bond (S–S) | Covalent single (biomolecular) | 240 | Cystine linkage in proteins |
| Amide (peptide) bond | Covalent resonance-stabilized | 350 | Peptide bond linking amino acids |
| Hydrogen bond | Noncovalent hydrogen bonding | 4-40 | Water dimer; DNA base pairing |
| Halogen bond | Noncovalent halogen bonding | 5-30 | Iodine···nitrogen interactions |
| Dipole–dipole interaction | Noncovalent electrostatic | 5-25 | Hydrogen chloride pairs |
| Ion–dipole interaction | Noncovalent ionic–polar | 20-200 | Na+ solvated by H2O |
| London dispersion (induced dipole) | Noncovalent dispersion | 0.05-40 | Noble gas condensation; hydrocarbons |
| π–π stacking | Noncovalent aromatic interaction | 2-12 | Stacked benzene rings |
| Cation–π interaction | Noncovalent electrostatic/π | 10-50 | Na+ interacting with benzene |
| Hydrophobic effect | Noncovalent entropic association | 1-10 | Protein folding, micelle formation |
| Salt bridge (ion pair) | Noncovalent ionic pair | 20-100 | Lysine–glutamate in proteins |
Images and Descriptions

Ionic bond
An electrostatic attraction between oppositely charged ions in salts and ionic solids. Strength depends on charge and distance; lattice energies can be hundreds to thousands of kJ/mol. Notable for forming high-melting, brittle crystals and conducting when molten or dissolved.

Metallic bond
A bond where valence electrons are delocalized across a lattice of metal cations, giving a “sea of electrons.” Cohesive energies vary widely; explains conductivity, malleability, and metallic luster in elemental metals and alloys.

Nonpolar covalent
An even sharing of electron pairs between atoms with similar electronegativities. Bond strengths range by atom types; common in diatomic gases and hydrocarbon frameworks. Notable for low polarity and poor solubility in water.

Polar covalent
Unequal sharing of electrons between atoms of differing electronegativity, producing partial charges. Energies overlap with covalent bonds; polarity leads to dipoles that influence solubility, reactivity, and intermolecular interactions like hydrogen bonding.

Single covalent
A single two-electron bond (usually a sigma bond) connecting two atoms. Typical energies depend on elements involved; common backbone bond in organic molecules. Notable for rotational freedom around the bond axis.

Double covalent
A bond composed of one sigma and one pi overlap, stronger and shorter than a single bond. Found in alkenes and carbonyl-containing compounds; restricts rotation and affects reactivity and conjugation.

Triple covalent
One sigma plus two pi bonds between atoms, producing short, very strong bonds. Typical in alkynes and N≡N gas; notable for high bond energy and linear geometry around the bond.

Coordinate (dative) bond
A covalent bond where both electrons come from the same atom (donor to acceptor). Energies similar to ordinary covalent bonds; common in metal complexes, Lewis acid–base adducts, and enzyme active sites.

Sigma bond (σ)
A covalent bond formed by head-on orbital overlap between atoms; usually the strongest component of covalent bonds. Sigma bonds determine primary connectivity and allow or restrict rotation depending on bonded partners.

Pi bond (π)
A covalent bond formed by side-by-side overlap of p orbitals, weaker than sigma bonds and contributing to double/triple bonds. Pi bonds lower bond rotation and participate in conjugation and aromaticity.

Aromatic bond (delocalized)
Delocalized pi bonding across a cyclic system producing enhanced stability (aromaticity). Bonds are intermediate between single and double, giving equalized lengths and unique chemical behavior like resonance stabilization.

Disulfide bond (S–S)
A covalent bond between two sulfur atoms, important in stabilizing protein tertiary and quaternary structures. Moderate strength; formed by oxidation of two thiols and notable for influencing stability and folding of proteins.
Amide (peptide) bond
A covalent bond between amino and carbonyl groups with partial double-bond character due to resonance, making it relatively strong and planar. Central to proteins and notable for restricted rotation and stability of peptide backbones.

Hydrogen bond
A directional attraction where a hydrogen attached to an electronegative atom interacts with another lone pair. Common in water, biological macromolecules, and polymers. Strength varies widely; crucial for structure and recognition.

Halogen bond
An attractive interaction where an electrophilic region on a halogen interacts with a nucleophile (lone pair or π system). Weaker than covalent bonds but directional; used in crystal engineering and molecular recognition.

Dipole–dipole interaction
Attractions between permanent molecular dipoles aligning opposite charges. Moderate strength and short range; influences boiling points and solubility of polar molecules and contributes to molecular ordering.

Ion–dipole interaction
Electrostatic attraction between an ion and a polar molecule’s dipole; can be quite strong for small, highly charged ions. Critical in solvation, dissolution of salts, and biological ion binding sites.

London dispersion (induced dipole)
Weak, universal attractions from instantaneous induced dipoles; strength increases with polarizability and contact area. Dominant in nonpolar molecules and large systems; can cumulatively be significant in macromolecules and solids.

π–π stacking
Attractive interactions between aromatic π systems driven by quadrupole and dispersion forces. Important in DNA base stacking, organic electronics, and supramolecular assemblies; strength depends on geometry and substituents.

Cation–π interaction
An attraction between a cation and the electron-rich face of a π system. Found in protein binding sites and receptors; relatively strong among noncovalent forces and important for molecular recognition.

Hydrophobic effect
An entropy-driven association of nonpolar surfaces in water, not a specific bond but a driving force for aggregation. Effective free-energy contribution depends on size and solvent; central to membrane formation and protein structure.

Salt bridge (ion pair)
A combined electrostatic and hydrogen-bonding interaction between oppositely charged groups (ion pair). Common in proteins and ionic complexes; strength depends on environment and distance, stabilizing tertiary structures.